Abstract
A large number of genomic and post-genomic studies in this century have been conducted to provide insight into the molecular mechanisms of gastrointestinal carcinogenesis and precision medicine. However, the incidence and mortality associated with gastrointestinal cancer have been increasing in the higher age range. Here, this review presents preemptive medicine to delay or prevent the onset of gastrointestinal cancer through an intervention based on molecular biomarkers, including an individual’s DNA, mtDNA, and microRNAs obtained by liquid biopsy.
Introduction
Striking advances in molecular dissection of gastrointestinal carcinogenesis in the last century have revealed that multiple genetic and epigenetic alterations in oncogenes, tumor-suppressor genes, and genetic instability, as well as telomerase activation, are implicated in the multistep process of gastrointestinal carcinogenesis [1,2]. Regarding gastric carcinogenesis, separate combinations of these alterations are seen in the different histological types of gastric cancer, indicating that intestinal and diffuse-type gastric cancers have distinct genetic pathways [3]. In addition, cancer-stromal interplay through the EGF receptor system’s network results in the invasion, progression, and metastasis in the setting of the gastric cancer micro-environment [4].
Since the beginning of the 21st century, a large number of genomic and post-genomic studies have been conducted to provide insight into the molecular mechanisms of carcinogenesis and precision medicine [5]. In fact, some of the oncogene mutations have been successfully targeted for molecular therapy of gastrointestinal cancer. Even so, the incidence and mortality associated with gastrointestinal cancer have been increasing remarkably in the higher age range in Japan [6].
Here, preemptive medicine is proposed to provide a promising approach to reduce the development of gastrointestinal cancer.
Concept and Strategy of Preemptive Medicine
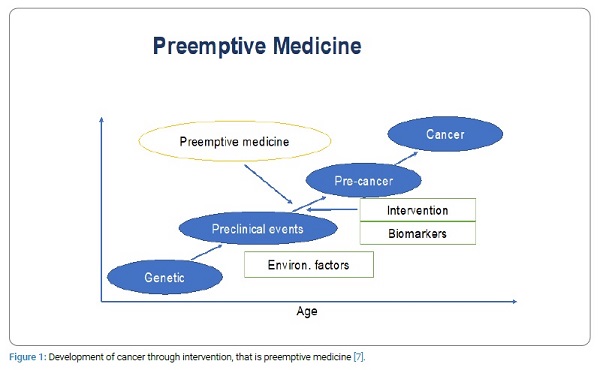
Preemptive medicine is a new concept of personalized preventive medicine to delay or prevent the onset of diseases through the intervention based on biomarkers including an individual’s DNA, RNA, protein and clinical imaging [7]. There are two levels in preemptive medicine. Namely, level 1 is inhibiting progression of the basic disease process such as chronic gastritis infected with Helicobacter pylori (Hp). Level 2 is diagnosing high risk group to prevent serious events such as hereditary and familial cancers.
Genetic and environmental factors bring about preclinical events which have high possibility to develop into cancer. If these preclinical events can be detected by biomarkers, we are able to delay or prevent the development of cancer through intervention, that is preemptive medicine (Figure 1).
The incidence rate of gastric adenocarcinoma has decreased in Japan, whereas the incidence rates of both proximal gastric and gastroesophageal junction adenocarcinoma have increased in both the United States and Europe [8]. Chronic infection with H. pylori, which is the strongest risk factor for developing gastric adenocarcinoma, induces alterations in gastric microbiota, while H. pylori eradication brings about an increase in bacterial diversity and restoration of the relative abundance of other bacteria [9].
On the other hand, recent studies of the gastrointestinal microbiome have identified alterations in the microbiota of the stomach, esophagus, and colon that increase the cancer risk [10]. In fact, the study of mucosal microbiome dysbiosis in gastric carcinogenesis revealed differences in bacterial interaction across stages of gastric carcinogenesis, suggesting that oral microbes P. stomatis, S. exigua, P. micra, S. anginosus and D. pneumosintes might play key roles in gastric carcinogenesis [11]. Microbial dysbiosis is also different between colonic adenoma and colorectal cancer patients [12]. Patients with adenoma showed higher relative abundances of Blautia, Ruminococcuss, Clostridium, and Lachnospiraceae compared to patients with colorectal cancer, while patients with colorectal cancer had higher relative abundances of Fusobacterium, Bacteroides, Phascolarctobacterium, and Porphyromonas compared to patients with adenoma [12].
Biomarkers for Preemptive Medicine
Although nuclear genetic and epigenetic alterations are the cornerstones of biomarkers for cancer, mitochondrial cellular content and mutations are also emerging as new molecular biomarkers [13,14]. Recently, the Genotype Tissue Expression (GTEx) project described that the cancer risk allele (G) of rs10826175 was associated with increased human mitochondrial transcription factor (TFAM) mRNA in various tissues including the esophagus and the stomach, suggesting that the upregulation of TFAM was associated with increased cancer susceptibility [15,16]. Moreover, a case control study entitled as “Identification of sequence polymorphisms in the mitochondrial D-loop as a risk factor for gastric cancer” reported that the frequent alleles of nucleotide 73G/A, 235A/G. 309C/C insert, 324C/G, 16,519C/T were significantly associated with an increased risk for gastric cancer [17]. The same research group confirmed that D-loop SNP of 16519C was associated with higher 8-hydrooxy- 2’-deoxyguanisine levels initiating carcinogenesis [18]. D-loop mutations have been also found in 40% of colorectal cancer [13]. Interestingly, circulating levels of mtDNA constitutes a noninvasive biomarker for the early detection of gastric cancer [19]. Patients with low mtDNA levels are likely to present precancerous or gastric cancer, whereas patients with high mtDNA levels are more likely to already present gastric cancer [19].
Exosomal miroRNA (mirRNA), which acts as a key player for cancer-stromal interaction, is a good candidate for biomarkers and therapeutic targets [20]. In fact, combination of four serum exosomal miRNAs including miR-92b-39,146b-5p,-9-5p, and let- 7g-5p is useful as novel non-invasive biomarkers for gastric cancer [21]. As regards colorectal adenoma, Canaan et al. reported that an eight miRNAs panel including miR-532-3p, miR-331, miR-195, miR-17, miR-142-3p, miR-15b, miR-532, and miR-652 efficiently distinguished patients with precancerous lesions form healthy control [22]. Moreover, methylated miRNAs have also recognized as cancer biomarker. For example, methylation of miR-17-5p, miR- 21-5p, miR-200-3p and let-7a-5p were significantly increased in gastrointestinal cancer and the methylation level of miR-17-5p in serum distinguished early pancreas cancer patients from healthy controls with extremely high sensitivity and specificity [23]. More excitingly, the combination of circulating miR and insoforms of microRNA (isomiR) detected by recent next generation sequencer acts as novel biomarkers for esophageal squamous cell carcinoma (ESCC) [24]. One mature miR-30a-5p and two isomR of mirR-574-3p and miR-205-5p in serum were significantly higher in ESCC patients than sex matched healthy control [24].
Regarding miRNA-based therapy, tumor suppressor miR-34a has been shown to down-regulate the expression of over 30 oncogenes including MET, MEK1, PDGFR-α, CDK4/6, BCL2, WINT1/3. NOTCH, CD44 as well as PD-L1. MRX34, a liposome-encapsulated miR- 34a mimic seems to be a potential first-in class miRNA therapy not only for cancer but also for precancerous lesions [25].
The liquid biopsy detecting circulating tumor DNA (ctDNA), mtDNA and microRNAs in the blood can be used to identify a high risk for cancer or at a very early and invisible stage. In fact, among the various biomarkers of ctDNA for colorectal cancer patients (stages 0-1), SEPT9 methylation is a promising biomarker for population screening [26]. In the future, liquid biopsy is a powerful tool to carry out the preemptive medicine, providing us with personalized cancer prevention. Finally, the longitudinal analysis of ctDNA is very useful to do residual disease detection and risk-stratified postoperative management of colorectal cancer [27].
Conclusion
Preemptive treatment is the ultimate goal of medicine but requires the analysis of large–scale whole genome sequencing (WGS) data integrated with RNA-Seq, epigenomics, immuno-genomic and clinic-pathological information. In addition, artificial intelligence (AI) may be needed for the effective implementation of preemptive medicine in the near future.
Acknowledgement
This review was presented at the Anniversary Lecture of the 30th Annual Meeting of the Japanese Society for Gastroenterological Carcinogenesis to be held on Nov.7,2019 in Yokohama.
Keywords
Gastrointestinal cancer; Preemptive medicine; Genome analysis; GI carcinogenesis; Biomarkers; Liquid biopsy
Cite this article
Tahara E. A new perspective for gastrointestinal cancer research-from genome analysis to preemptive medicine. Clin Oncol J. 2020;1(3):1–4.
Copyright
© 2020 Eiichi Tahara. This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International License (CC BY-4.0).

